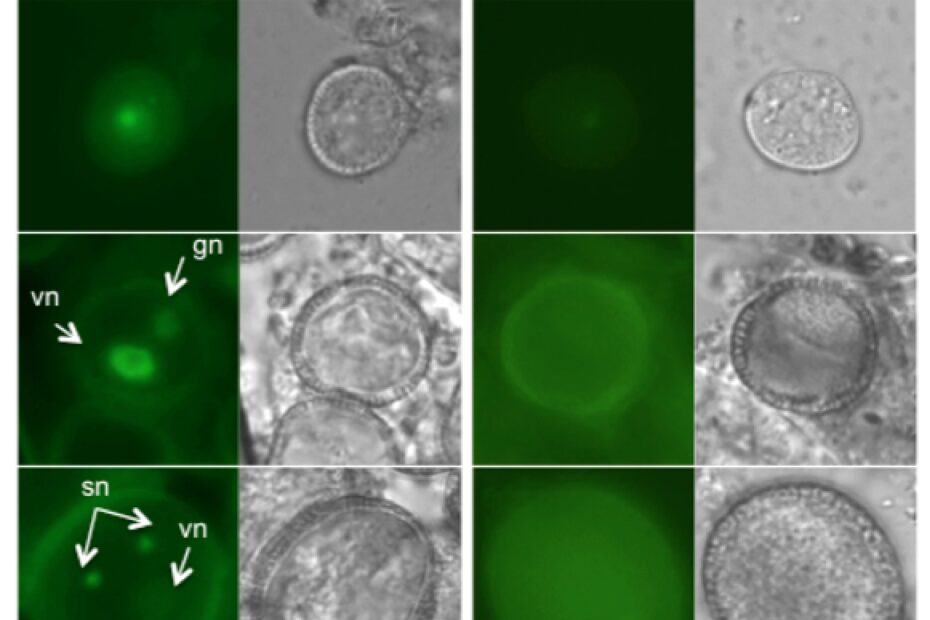
Abstract: Centromeres define the chromosomal position where kinetochores form to link the chromosome to microtubules during mitosis and meiosis. Centromere identity is determined by incorporation of a specific histone H3 variant termed CenH3. As for other histones, escort and deposition of CenH3 must be ensured by histone chaperones, which handle the non-nucleosomal CenH3 pool and replenish CenH3 chromatin in dividing cells. Here, we show that the Arabidopsis orthologue of the mammalian NUCLEAR AUTOANTIGENIC SPERM PROTEIN (NASP) and S. pombe histone chaperone Sim3 is a soluble nuclear protein that binds the histone variant CenH3 and affects its abundance at the centromeres. NASPSIM3 is co-expressed with Arabidopsis CenH3 in dividing cells and binds directly to both the N-terminal tail and the histone fold domain of non-nucleosomal CenH3. Reduced NASPSIM3expression negatively affects CenH3 deposition, identifying NASPSIM3 as a CenH3 histone chaperone.
Keywords: Arabidopsis thaliana,centromere, kinetochore, CenH3, histone chaperone, NASPSIM3
Samuel Le Goff1,#, BurcuNur Keçeli2,#, Hana Jerabkova3, Stefan Heckmann4, Twan Rutten4, Sylviane Cotterell1, Veit Schubert4, Elisabeth Roitinger5-7, Karl Mechtler5-7, F. Christopher H. Franklin8, Christophe Tatout1, Andreas Houben4, Danny Geelen2, Aline V. Probst1;*, and Inna Lermontova4,9;*
1 GReD, Université Clermont Auvergne, CNRS, INSERM, BP 38, 63001 Clermont–Ferrand, France
2 Department of Plants and Crops, unit HortiCell, Faculty of Bioscience Engineering, Ghent University, Coupure links, 653, 9000 Ghent, Belgium
3 The Czech Academy of Sciences, Institute of Experimental Botany (IEB), Centre of the Region Haná for Biotechnological and Agricultural Research, Slechtitelu 31, 783 71 Olomouc, Czech Republic
4 Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, Corrensstrasse 3, D-06466 Seeland, Germany
5 Institute of Molecular Pathology (IMP), Vienna BioCenter (VBC), Vienna 1030, Austria
6 Institute of Molecular Biotechnology (IMBA), Austrian Academy of Sciences, Vienna BioCenter (VBC), Vienna 1030, Austria
7 Gregor Mendel Institute (GMI), Austrian Academy of Sciences, Vienna BioCenter (VBC), Vienna 1030, Austria
8 School of Biosciences, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK9 Mendel Centre for Plant Genomics and Proteomics, CEITEC, Masaryk University, Brno CZ-62500, Czech Republic
# These authors contributed equally to this work.
*Authors for correspondence: lermonto@ipk-gatersleben.de & aline.probst@uca.fr Phone: ++49/39482 5570/ ++33/473407401Fax: ++49/39482 5137 ++33/473407777